Hello BioFreakers,
Today in the GGS LIVE section we will cover how to generate rabbit polyclonal antibody:)
Method: Established methodology for antibody generation is an essential tool to study cellular functions of the protein of interest.
About: Antibodies are generated by injection of the antigen into host animal (mouse, rat, rabbit etc) and subsequent isolation of antibodies from the animal serum (for instance from rabbit serum).
What: Generation of antibody against protein X.
There are many ways to generate antibodies but in our study case we are going to cover a protocol for generation of rabbit polyclonal antibody [monoclonal (recognise single epitope), polyclonal (recognises multiple epitopes)]. Ok, so lets see what are the main steps and tools here:
1. Antigen - design and generation - may be performed by you or company.
2. Animals - injection and animals maintanance - usually company
3. Validation of antibody - isolation and purification of antigen specific antibodies - you or company
In our study case, we will generate a polyclonal antibody against the protein of interest (size 25kDa). Because our protein is relatively small we can use its full sequence for rabbit immungenisation. In case of larger proteins (they still could be used as a whole, however purification of a big protein may be a limiting factor) a small peptide, usually from either N- or C-terminus of the protein is selected as an antigen. Please have a look at this animation showing an immune system response to antigens (Biology Animation -The Activatioof the Humoral and Cell-Mediated Pathways). This animation shows how antibodies are produced by the immune system.
Antigen generation
First we need to clone the Protein X DNA sequence into an appropriate vector that will allow us to express and purify the Protein X. In our study case we will use a GST tag for that purpose and we are going to purify it from E.coli cells. Cloning, expression and purification of tagged proteins have been previously described within different posts (just click GGS LIVE - PCR, -Cloning, -Protein tagging, -Expression and -Purification). Once we have the antigen purified (please see below):
we can contact the company that will generate the antibody for us. Before the immunogenisation we should check several pre-immune sera (serum from rabbits before the immunogenisation) to see if they are able to react with the proteins of a similar size as our protein of interest (important when antibody is going to be used primarily for immunoblotting aka Western blotting). Such a test by immunoblotting is shown below (click here for GGS LIVE - Immunoblotting tutorial):
As you can see, in our study case the test with pre-immune sera showed that all four sera react with proteins in lysates from human cells and detect a band of 75kDa. Moreover, three of them strongly recognise a band of molecular size of approximately 50kDa (sera 1, 3 and 4) and serum 2 picks up an extra band around 27kDa. Additionally, sera 3 and 4 recognise weakly two additional bands (size ~20kDa and ~30kDa for serum 3, ~18kDa and ~35kDa for serum 4). This analysis allow us to pick up a serum (which corresponds to an animal that will be injected with the antigen) that does not recognise a band of a similar size as our protein of interest before immunogenisation. As you can see sera 1 and 4 are the only ones that do not pick up any bands in the area where our protein of interest may be migrating in the gel (between 20 and 35 kDa). Usually, the same antigen is injected into two animals to increase the chance of generating a working antibody, therefore we are going to choose rabbit 1 and 4 for our immunigenisation protocol.
After antigen generation and choosing the appropriate animals for immunogenisation, we send our antigen to the company which is going to inject it into rabbits and then provide us with immunogenised sera. This usually takes 4-12 weeks depending on the protocol used. During this time company usually provides three sera (1st, 2nd and final). These are then tested to see if they recognise the protein of interest using immunoblotting or any other immuno-technique (such as immunofluorescence or ELISA). Purified protein of interest (antigen - in our case Protein X) and cell lysates are used for these purposes. In the ideal situation the final serum (or even better 1st or 2nd one) recognises the protein of interest in the cell lysates without prior purification of the antigen specific antibodies from the serum. However, commonly the final sera recognises the antigen (a very good start and a must do in this case:)) but fail to recognise the protein of interest in the cell lysates. It also happens that the final serum is "dirty" and the immunoblots have a lot of non-specific, background signal that interferes with the detection of our protein (see below).
As you can see serum before and after the purification can easily detect the antigen but only the purified antibody can recognise the protein X in the cell lysates. This is very often due to the low concentration of the antigen specific antibodies in the serum compared to other antibodies. Therefore, the stronger the specific signal we want to obtain the stronger the background. This way it is very hard to get a strong enough signal (nice band on the right hand film above) for the protein X in the cell lysates that would be visible over the background noise. However, as you can clearly see purification of the antibody solved the problem (see here GGS LIVE - Antibody purification against the antigen).
Next step is to test if the antibody can be used in other applications such as immunofluorescence (GGS LIVE - Immunofluorescence) and immunoprecipitation (GGS LIVE - Immunoprecipitation).
I hope you enjoyed.
GGS TEAM
Showing posts with label Biochemistry Methods. Show all posts
Showing posts with label Biochemistry Methods. Show all posts
Saturday, 2 May 2015
Tuesday, 28 April 2015
GGS LIVE - Bacterial Transformation
Welcome Biofreakers,
Today, GGS TEAM is happy to present the bacterial tranformation technique.
Method: Transformation of E.coli cells with DNA plasmid.
About: This technique allows for introduction of foreign DNA into bacteria cells in order to for example propagate the DNA or express a protein of interest.
What: Transformation of E.coli strain TOP10 with pEGFPN1 plasmid containing MCM2 cDNA (pEGFPN1-MCM2).
Transforming plasmid DNA into bacteria cells is fast and easy. Of course we need bacteria cells and for that purpose different E. coli strains (such as TOP10, DH5alpha or others) are usually used. These cells are previously prepared in order to receive the DNA in the process that makes bacteria cells "competent". Such competency is nothing more than making the bacterial cell wall transiently penetrable (pores in the bacterial cell wall). Such condition of bacterial "coatings" allows for the take up of the DNA. Ok, lets start then. On the Figure below you can see an outline of the bacterial transformation process.
First competent bacteria are prepared using variety of different methods. These cells are then snap frozen using liquid nitrogen and stored at -80C to -150C to preserve to competent state. Once the cells are needed they are placed onto ice to defrost. On ice the cells are still competent but their ability to uptake DNA decreases with time and with increasing the temperature. Next, cells are mixed with DNA, in our case the DNA is the pEGFPN1-MCM2 plasmid. If the DNA that is used for transformation is a pure plasmid we do not need to use a lot but if the DNA we are using is for example DNA ligation reaction, we should use as much as possible to increase a chance of getting our DNA into the cells. After addition of the DNA the bacteria-DNA mix is incubated on ice for 20-30min in order to create "bacteria-DNA complexes". Next, the reaction is transfered to 42C (usually a water bath or hot plate) and incubated for a short time such as 90s. This step called heat shock opens previously introduced pores (these appeared when cells were made competent) and allows up-taking DNA that was previously in the contact with bacterial cell wall.After the heat shock there is a time for cold shock at 4C to close the pores and traps the DNA inside bacterial cells. After the "shocks" cells are allowed to recover by addition of fresh media and incubation at 37C shaking for approximately two cell cycles which in case of E coli is approximately 40min. Cells are then seeded onto plates containing appropriate antibiotic, in our case it is the Kanamycin and plates are the placed in the 37C incubator; and incubated over night. The amount of the cells seeded also depends on the DNA that was transformed. As previously mentioned, if the DNA was a pure plasmid we can seed very little (approximately 5-10% of the transformation) but if the DNA came from the ligation reaction we can seed all the transformation to be sure we get colonies back Next day, the colonies appear on the plates. Single colony is then picked up and expanded as a culture. Such culture can be then used to isolate bigger quantities of the DNA which later can be used for other purposes.
I hope you enjoyed my come back:)
GGS TEAM
Today, GGS TEAM is happy to present the bacterial tranformation technique.
Method: Transformation of E.coli cells with DNA plasmid.
About: This technique allows for introduction of foreign DNA into bacteria cells in order to for example propagate the DNA or express a protein of interest.
What: Transformation of E.coli strain TOP10 with pEGFPN1 plasmid containing MCM2 cDNA (pEGFPN1-MCM2).
Transforming plasmid DNA into bacteria cells is fast and easy. Of course we need bacteria cells and for that purpose different E. coli strains (such as TOP10, DH5alpha or others) are usually used. These cells are previously prepared in order to receive the DNA in the process that makes bacteria cells "competent". Such competency is nothing more than making the bacterial cell wall transiently penetrable (pores in the bacterial cell wall). Such condition of bacterial "coatings" allows for the take up of the DNA. Ok, lets start then. On the Figure below you can see an outline of the bacterial transformation process.
First competent bacteria are prepared using variety of different methods. These cells are then snap frozen using liquid nitrogen and stored at -80C to -150C to preserve to competent state. Once the cells are needed they are placed onto ice to defrost. On ice the cells are still competent but their ability to uptake DNA decreases with time and with increasing the temperature. Next, cells are mixed with DNA, in our case the DNA is the pEGFPN1-MCM2 plasmid. If the DNA that is used for transformation is a pure plasmid we do not need to use a lot but if the DNA we are using is for example DNA ligation reaction, we should use as much as possible to increase a chance of getting our DNA into the cells. After addition of the DNA the bacteria-DNA mix is incubated on ice for 20-30min in order to create "bacteria-DNA complexes". Next, the reaction is transfered to 42C (usually a water bath or hot plate) and incubated for a short time such as 90s. This step called heat shock opens previously introduced pores (these appeared when cells were made competent) and allows up-taking DNA that was previously in the contact with bacterial cell wall.After the heat shock there is a time for cold shock at 4C to close the pores and traps the DNA inside bacterial cells. After the "shocks" cells are allowed to recover by addition of fresh media and incubation at 37C shaking for approximately two cell cycles which in case of E coli is approximately 40min. Cells are then seeded onto plates containing appropriate antibiotic, in our case it is the Kanamycin and plates are the placed in the 37C incubator; and incubated over night. The amount of the cells seeded also depends on the DNA that was transformed. As previously mentioned, if the DNA was a pure plasmid we can seed very little (approximately 5-10% of the transformation) but if the DNA came from the ligation reaction we can seed all the transformation to be sure we get colonies back Next day, the colonies appear on the plates. Single colony is then picked up and expanded as a culture. Such culture can be then used to isolate bigger quantities of the DNA which later can be used for other purposes.
I hope you enjoyed my come back:)
GGS TEAM
Saturday, 25 August 2012
GGS LIVE - Protein expression in vertebrate cells
Hello Biofreakers,
I am back BAYBE!!
Today we will cover a topic about expression of protein in vertebrate cells.
Method: Expression of protein X in human osteosarcoma cells (U2OS).
About: Protein expression allows for either purification or study of its cellular function through for example its localisation etc.
What: Expression of RFP-tagged-(Red Fluorescent Protein)-Protein X in U2OS cells to study its localisation.
To express a protein of interest in vertebrate cells we need to have its cDNA sequence. To get the target cDNA we need to clone it from either cDNA library (optionally isolate the cDNA from such library) or from mRNA. When cDNA sequence is ready it is then cloned into a vector that allows expression of that cDNA in vertebrate cells (please see this post GGS LIVE - Making a fusion protein for more details on how to create fusion proteins). Different vectors are available for expression of cDNAs in vertebrate cells. When vector containing cDNA is generated it has to be transfected into cells. This can be achieved by electroporation, chemical reagent such as lipid based transporters and others.
In our study case the U2OS cells will were transfected with lipid based reagent (lipofectamine). In order to detect the protein of interest its expression was monitored by fluorescence microscopy (please see this post for more details GGS LIVE - Immunofluorescence) as shown on Figure below.

As you can see on the Figure, not transfected cell show very little or no red fluorescence indicating that there is no RFP-Protein X expressed in these cells. On the contrary the cells transfected with the cDNA coding RFP-Protein X fusion show red fluorescence. Moreover, the red fluorescence co-localises with the blue signal that comes from DNA (DNA was stained with DAPI). This suggests that the RFP-Protein X can be mainly found in the nucleus. Therefore, Protein-X is very likely to be a nuclear protein (to test that we could use technique described here GGS LIVE - Cell Fractionation). To confirm that we indeed expressed the RFP-protein X in U2OS cells, protein extract were prepared from these cells after transfection and analysed by immunoblotting (for more details on this technique please see here GGS LIVE - Western Blotting).
As you can see on the Figure below, the immunoblotting showed that RFP-Proiten X fusion can be detected exclusively in the cells that were transfected with the DNA coding for RFP-Protein X (antibody against the RFP was used to detect RFP-Protein X fusion). Detection of Actin protein was used here as loading control showing that the lane NT contains proteins but indeed does not have Protein X.
Moreover, the protein that we wish to express may not contain tag or be fused to a different type of tag (eg. non-fluorecent tag) that allows for example, to purify and study the biochemistry of the Protein X or to identify other proteins bound to protein X. This can be achieved by immnuprecipitation of the Protein X and analysis of the protein complexes associated with Protein X (for immunoprecipitation thechnique go here Biochemistry Methods - Immunoprecipitation).
As you can see the there is many ways that we could use protein expression as a tool to answer our scientific questions.
I hope you enjoyed it:)
Cheers,
GGS TEAM
I am back BAYBE!!
Today we will cover a topic about expression of protein in vertebrate cells.
Method: Expression of protein X in human osteosarcoma cells (U2OS).
About: Protein expression allows for either purification or study of its cellular function through for example its localisation etc.
What: Expression of RFP-tagged-(Red Fluorescent Protein)-Protein X in U2OS cells to study its localisation.
To express a protein of interest in vertebrate cells we need to have its cDNA sequence. To get the target cDNA we need to clone it from either cDNA library (optionally isolate the cDNA from such library) or from mRNA. When cDNA sequence is ready it is then cloned into a vector that allows expression of that cDNA in vertebrate cells (please see this post GGS LIVE - Making a fusion protein for more details on how to create fusion proteins). Different vectors are available for expression of cDNAs in vertebrate cells. When vector containing cDNA is generated it has to be transfected into cells. This can be achieved by electroporation, chemical reagent such as lipid based transporters and others.
In our study case the U2OS cells will were transfected with lipid based reagent (lipofectamine). In order to detect the protein of interest its expression was monitored by fluorescence microscopy (please see this post for more details GGS LIVE - Immunofluorescence) as shown on Figure below.

As you can see on the Figure below, the immunoblotting showed that RFP-Proiten X fusion can be detected exclusively in the cells that were transfected with the DNA coding for RFP-Protein X (antibody against the RFP was used to detect RFP-Protein X fusion). Detection of Actin protein was used here as loading control showing that the lane NT contains proteins but indeed does not have Protein X.
The biggest issue when expressing the protein of interest in the vertebrate cells is the transfection efficiency. The highest transfection efficiencies are obtained when the specific viruses are used to deliver the DNA into host cells (usually more thatn 90%). Unfortunately, I do not have any experience with this approach, therefore it will not be discussed further here. However, to ensure that as much cells as possible express our protein of interest we can optimise our transfection conditions by testing different ratios of the transfection reagent to DNA. For an example, we can keep the amount of the DNA constant and change the volume of transfection reagent as shown in the table and figure below:
In this case I used Fusion of Green Fluorescent Protein (GFP) with Protein X as my marker. The DNA labelling with DAPI shows the total number of the cells. The number of the cells showing Green Fluorescence by the total number of the cells gives us the transfection efficiency. As you can see when increasing amount of transfection reagent was used the transfection efficiency rised.
Moreover, the protein that we wish to express may not contain tag or be fused to a different type of tag (eg. non-fluorecent tag) that allows for example, to purify and study the biochemistry of the Protein X or to identify other proteins bound to protein X. This can be achieved by immnuprecipitation of the Protein X and analysis of the protein complexes associated with Protein X (for immunoprecipitation thechnique go here Biochemistry Methods - Immunoprecipitation).
As you can see the there is many ways that we could use protein expression as a tool to answer our scientific questions.
I hope you enjoyed it:)
Cheers,
GGS TEAM
Monday, 13 June 2011
GGS LIVE - Site Directed Mutagenesis
Hello BioFreakers!!
Today in the GGS - LIVE section the Site Directed Mutagenesis technique. Wanna see how it is done? Lets roll then.
Method: Site Directed Mutagenesis (SDM).
About: Allows for introduction of point mutations in DNA and thus sometimes in protein sequence.
What: Introduction of STOP codon in order to generate C-terminal deletion in protein X.
Site directed mutagenesis is performed on circular DNA substrate. In our study case, the cDNA of protein X will be inserted into GST vector. Cloning of the protein X was performed analogously to study case described in a different post (click here GGS LIVE - Makinga fusion protein).
To mutate protein X cDNA sequence we will need a set of primers that will be used in PCR reaction (GGS LIVE - Polymerase chain reaction (PCR)). SDM primers overlap with the target sequence, whereas regular PCR primers flank the target sequence (see cartoon below).
As you can see from the gel electroporesis analysis, desired product is present in both DpnI treated and untreated samples (R and R+D). Template +/- DpnI was used as a negative control. Additionally, amplification of 0.3kb fragment served as template control.
You probably, wonder why SDM product and template are not observed as single band. This is due to different conformations of these plasmids. Template is supercoiled, therefore more packed and travels through gel faster and thus appears smaller on the gel. SDM product is not and therefore migrates slower.
The R+D sample is then used to transform bacterial cells. In this process specific E.coli cells (for example Top10) uptake R+D plasmid and replicate it. This allows obtaining workable amounts of DNA, which is then send for sequencing. Several different E.coli clones are tested and when sequencing confirms mutation of pPLAS + X cDNA, DNA from correct clone is used to transform yet another specifc E.coli strain (for example BL21 pLysS).
We then express X and X-truncation proteins using these BL21 cells (see figure below). For the protein expression in E.coli tutorial, please go here GGS LIVE - Protein expression in E.coli.
Today in the GGS - LIVE section the Site Directed Mutagenesis technique. Wanna see how it is done? Lets roll then.
Method: Site Directed Mutagenesis (SDM).
About: Allows for introduction of point mutations in DNA and thus sometimes in protein sequence.
What: Introduction of STOP codon in order to generate C-terminal deletion in protein X.
Site directed mutagenesis is performed on circular DNA substrate. In our study case, the cDNA of protein X will be inserted into GST vector. Cloning of the protein X was performed analogously to study case described in a different post (click here GGS LIVE - Makinga fusion protein).
To mutate protein X cDNA sequence we will need a set of primers that will be used in PCR reaction (GGS LIVE - Polymerase chain reaction (PCR)). SDM primers overlap with the target sequence, whereas regular PCR primers flank the target sequence (see cartoon below).
The idea behind SDM is that each primer will mutate a single DNA strand of the pPLAS + X cDNA plasmid giving a product of the mutated pPLAS + X cDNA. Naturally, the template will be also present in the final mixture. Tamplate is removed by DpnI, a restriction endonuclease that digest modified DNA (DNA methylation). Such modification of the DNA occurs within bacterial cells, therefore SDM product will not wear it, as it was formed in vitro. After PCR reaction and overnight digestion with DpnI, SDM reaction is analysed by agarose gel electrophoresis (see figure below).
You probably, wonder why SDM product and template are not observed as single band. This is due to different conformations of these plasmids. Template is supercoiled, therefore more packed and travels through gel faster and thus appears smaller on the gel. SDM product is not and therefore migrates slower.
The R+D sample is then used to transform bacterial cells. In this process specific E.coli cells (for example Top10) uptake R+D plasmid and replicate it. This allows obtaining workable amounts of DNA, which is then send for sequencing. Several different E.coli clones are tested and when sequencing confirms mutation of pPLAS + X cDNA, DNA from correct clone is used to transform yet another specifc E.coli strain (for example BL21 pLysS).
We then express X and X-truncation proteins using these BL21 cells (see figure below). For the protein expression in E.coli tutorial, please go here GGS LIVE - Protein expression in E.coli.
As you can see, this way we can generate a mutant protein KA-CHING.
I hope you enjoyed it.
Maciek
GGSTEAM
Thursday, 9 June 2011
GGS LIVE - Protein purification from E.coli
Yo Yo Biofreakers,
Today we are going to have closer look at the recombinant protein purification from bacterial cells.
Method: Purification of recombinant protein.
About: Having an optimised protocol for protein purification is an essential tool to study properties of the protein of interest.
What: Purification of GST-tagged chicken protein previously expressed in E.coli.
Before we start we should first have a look at what the GST tag is. GST (glutathione S-transferase) is an enzyme that transfer glutathione (GSH) via slufhydryl group to differernt type of substrates (lipids, xenobiotics). GST has a high affinity towards its substrate glutathione and this property of GST is utilised to purify GST-tagged proteins. In order to recover GST-tagged protein from the complicated mixture of proteins, the fusion protein is incubated with agarose beads coupled to glutathione (GSH-agarose, see picture below), what leads to efficient precipitation of GST-fusion protein.
Ok Vamos!! Expression of the GST-X protein was previously demonstrated in different post GGS LIVE - Protein Expression in E.coli.
After a succesfull expression of the protein of interest in E.coli, we can purify it using one of the many protocols available for GST protein fusion purification. In general such protocol consists of three major steps: cell lysis and solubilisation of the GST-fusion protein (see cartoon below), recovery of the fusion from the lysate and elution of the GST fusion.
As you can see on the cartoon above, first GST-X fusion is expressed in large amount (for example 250 ml to 1 l culture) under previously optimised conditions (for protein expression see post GGS LIVE - Protein expression in E.coli). After protein expression, E.coli cells are harvested by centrifugation, superatant is removed and cell pellet is resuspended in lysis buffer of choice. Usually such buffer should have pH of around 7.0 - 8.0 to facilitate efficient interaction between GST and its substrate glutathione, protease inhibitors (such as PMSF) to prevent protein degradation. Additionally, lysis buffer should contain component taht will help lyse the cells, such as lysosyme (enzyme that degrades bacterial cell wall) or detergent (which disrupts bacterial cell wall). Cells are usually lysed at 4C rocking or mildly shaking what increases lysis efficiency. Cell lysate is then sonicated to share bacterial DNA (DNA makes lysate viscous and hard to work with) and help to solubilise proteins by breaking up protein aggregates. In the next step, cell debri is removed by high speed centrifugation. And there we go we have a lysate ready for protein purification.
As mentioned earlier, in order to recover our GST-X protein we have to mix our lysate containg the fusion with glutathione agarose beads. First beads have to be prepared (see cartoon below).
Glutathione agarose beads are first washed with the lysis buffer in order to remove storage solution (usually ethanol, which can impede binding of GST to glutathione). Then beads are mixed with lysate containing GST-X fusion and incubated at 4C in order to bind GST fusion to the beads. After binding step, beads have to be washed in order to remove unbound GST-X fusion and unspecifically bound proteins.
At this stage of purification GST-X fusion should be clean and depending on the nature of furhter experiments that we want to perform, we can either elute the fusion with glutathione (excess of the glutathione will compete and displace GST-X protein from the beads) or cleave the GST tag and release protein X (see cartoon below).
When purification is finished we can analyse our experiment by separating protein sample taken at each step of the purification by SDS-PAGE and stain proteins in gel with Coomasie dye. The results from GST-X purification are shown on the picture below.
As you can see from the Coomasie stained gel, the expression of GST-X fusion was nicely induced (UI and I samples). We can also confirm that the GST-X was present in the starting material (lysate IN sample). After incubation of the lysate with beads, most of the GST-X bound to the resin what resulted in depletion of GST-X, as observed in unbound sample (UN). After beads wash, a single band of GST-X was detected on the beads, indicating high purifty of this sample. Elution of the GST-X with glutathione recovered fusion protein from beads. Additionally, alternative elution by GST cleavage resulted in appeareance of two bands: a free X protein and GST tag.
Hopefully, you got the picture how protein purification can be performed using a GST tag as a bait.
I hope u enjoyed it.
Cu SOON!
Maciek
GGSTEAM
Today we are going to have closer look at the recombinant protein purification from bacterial cells.
Method: Purification of recombinant protein.
About: Having an optimised protocol for protein purification is an essential tool to study properties of the protein of interest.
What: Purification of GST-tagged chicken protein previously expressed in E.coli.
Before we start we should first have a look at what the GST tag is. GST (glutathione S-transferase) is an enzyme that transfer glutathione (GSH) via slufhydryl group to differernt type of substrates (lipids, xenobiotics). GST has a high affinity towards its substrate glutathione and this property of GST is utilised to purify GST-tagged proteins. In order to recover GST-tagged protein from the complicated mixture of proteins, the fusion protein is incubated with agarose beads coupled to glutathione (GSH-agarose, see picture below), what leads to efficient precipitation of GST-fusion protein.
Ok Vamos!! Expression of the GST-X protein was previously demonstrated in different post GGS LIVE - Protein Expression in E.coli.
After a succesfull expression of the protein of interest in E.coli, we can purify it using one of the many protocols available for GST protein fusion purification. In general such protocol consists of three major steps: cell lysis and solubilisation of the GST-fusion protein (see cartoon below), recovery of the fusion from the lysate and elution of the GST fusion.
As you can see on the cartoon above, first GST-X fusion is expressed in large amount (for example 250 ml to 1 l culture) under previously optimised conditions (for protein expression see post GGS LIVE - Protein expression in E.coli). After protein expression, E.coli cells are harvested by centrifugation, superatant is removed and cell pellet is resuspended in lysis buffer of choice. Usually such buffer should have pH of around 7.0 - 8.0 to facilitate efficient interaction between GST and its substrate glutathione, protease inhibitors (such as PMSF) to prevent protein degradation. Additionally, lysis buffer should contain component taht will help lyse the cells, such as lysosyme (enzyme that degrades bacterial cell wall) or detergent (which disrupts bacterial cell wall). Cells are usually lysed at 4C rocking or mildly shaking what increases lysis efficiency. Cell lysate is then sonicated to share bacterial DNA (DNA makes lysate viscous and hard to work with) and help to solubilise proteins by breaking up protein aggregates. In the next step, cell debri is removed by high speed centrifugation. And there we go we have a lysate ready for protein purification.
As mentioned earlier, in order to recover our GST-X protein we have to mix our lysate containg the fusion with glutathione agarose beads. First beads have to be prepared (see cartoon below).
Glutathione agarose beads are first washed with the lysis buffer in order to remove storage solution (usually ethanol, which can impede binding of GST to glutathione). Then beads are mixed with lysate containing GST-X fusion and incubated at 4C in order to bind GST fusion to the beads. After binding step, beads have to be washed in order to remove unbound GST-X fusion and unspecifically bound proteins.
At this stage of purification GST-X fusion should be clean and depending on the nature of furhter experiments that we want to perform, we can either elute the fusion with glutathione (excess of the glutathione will compete and displace GST-X protein from the beads) or cleave the GST tag and release protein X (see cartoon below).
When purification is finished we can analyse our experiment by separating protein sample taken at each step of the purification by SDS-PAGE and stain proteins in gel with Coomasie dye. The results from GST-X purification are shown on the picture below.
As you can see from the Coomasie stained gel, the expression of GST-X fusion was nicely induced (UI and I samples). We can also confirm that the GST-X was present in the starting material (lysate IN sample). After incubation of the lysate with beads, most of the GST-X bound to the resin what resulted in depletion of GST-X, as observed in unbound sample (UN). After beads wash, a single band of GST-X was detected on the beads, indicating high purifty of this sample. Elution of the GST-X with glutathione recovered fusion protein from beads. Additionally, alternative elution by GST cleavage resulted in appeareance of two bands: a free X protein and GST tag.
Hopefully, you got the picture how protein purification can be performed using a GST tag as a bait.
I hope u enjoyed it.
Cu SOON!
Maciek
GGSTEAM
Monday, 14 February 2011
GGS LIVE - Protein expression in E.coli
Dear BioFreaker,
today in the GGS LIVE section we are going to look at the recombinant protein expression in E.coli.
Method: Protein expression.
About: Protein expression in E.coli is a widely applied technique, which allows for quick and robust production of the particular protein. Such protein can be then used in different applications, such as antibody production, in vitro enzymatic and binding assays, crystallisation etc.
What: Expression of the GST-tagged chicken protein in E.coli.
Ok, lets start. If we want to express any protein in the bacteria we have to first clone it (for more info about cloning please visit this post GGS LIVE cloning) into a plasmid which contains specific elements allowing for protein production within the prokaryotic bacteria cells. These plasmids utilise technology based on application of lac operon system (click here for more information about the lac operon system). We will not cover how this system works in this post, we will just go straight to the results:). The only thing you should know at this stage is that protein expression from the lac operon system is achieved by addition of IPTG compound. The IPTG turns on the protein production, which cannot happen without it.
If a technology for protein X purification is established then expression of such can be performed using native protein sequence. If such protocol is not available we can express our protein as a fusion. The tag which will be added to the protein will later allow for its rapid and robust purification. There are many different tags available for various applications, such as His, GST, MBP, CBP, S-tag, FLAG, Strep and many more. In our study case we will work with GST-tagged protein. If you would like to know how to tag a protein of interest, please look at this post GGS LIVE - Making a fusion protein.
After we cloned our DNA sequence of interest into a N-terminal GST tag containing vector, we have to transform E.coli strains with this plasmid and then find optimal conditions for expression of our fusion protein. This step has to be performed experimentally. The culture of bacterial cells is split into many batches where protein expression is performed at different conditions, such as IPTG concentration, temperature, time, different media composition etc. At the end of the experiment samples are separated by gel electrophopresis and proteins visualised by staining of the gel. Such gel staining, where protein expression at different temperatures was tested, is shown on the picture below:
Picture taken by Kliszczak M.
I have to mention that our protein of interest has size of 27 kDa and the GST tag is similarly big (28 kDa), what gives size of 55 kDa for the fusion. As you can observe on the gel above, increase of temperature during expression positively affects production of GST-X protein by E.coli cells. The 37C seem to be the best temperature for expression of fusion protein. In addition you can observe that GST-X has the predicted size of 55 kDa. After, establishment of perfect conditions, these can be used for production of our protein.
I hope you enjoyed. In the next post we will cover the purification of the GST fusion protein from E.coli.
Maciek GGS TEAM
today in the GGS LIVE section we are going to look at the recombinant protein expression in E.coli.
Method: Protein expression.
About: Protein expression in E.coli is a widely applied technique, which allows for quick and robust production of the particular protein. Such protein can be then used in different applications, such as antibody production, in vitro enzymatic and binding assays, crystallisation etc.
What: Expression of the GST-tagged chicken protein in E.coli.
Ok, lets start. If we want to express any protein in the bacteria we have to first clone it (for more info about cloning please visit this post GGS LIVE cloning) into a plasmid which contains specific elements allowing for protein production within the prokaryotic bacteria cells. These plasmids utilise technology based on application of lac operon system (click here for more information about the lac operon system). We will not cover how this system works in this post, we will just go straight to the results:). The only thing you should know at this stage is that protein expression from the lac operon system is achieved by addition of IPTG compound. The IPTG turns on the protein production, which cannot happen without it.
If a technology for protein X purification is established then expression of such can be performed using native protein sequence. If such protocol is not available we can express our protein as a fusion. The tag which will be added to the protein will later allow for its rapid and robust purification. There are many different tags available for various applications, such as His, GST, MBP, CBP, S-tag, FLAG, Strep and many more. In our study case we will work with GST-tagged protein. If you would like to know how to tag a protein of interest, please look at this post GGS LIVE - Making a fusion protein.
After we cloned our DNA sequence of interest into a N-terminal GST tag containing vector, we have to transform E.coli strains with this plasmid and then find optimal conditions for expression of our fusion protein. This step has to be performed experimentally. The culture of bacterial cells is split into many batches where protein expression is performed at different conditions, such as IPTG concentration, temperature, time, different media composition etc. At the end of the experiment samples are separated by gel electrophopresis and proteins visualised by staining of the gel. Such gel staining, where protein expression at different temperatures was tested, is shown on the picture below:
I have to mention that our protein of interest has size of 27 kDa and the GST tag is similarly big (28 kDa), what gives size of 55 kDa for the fusion. As you can observe on the gel above, increase of temperature during expression positively affects production of GST-X protein by E.coli cells. The 37C seem to be the best temperature for expression of fusion protein. In addition you can observe that GST-X has the predicted size of 55 kDa. After, establishment of perfect conditions, these can be used for production of our protein.
I hope you enjoyed. In the next post we will cover the purification of the GST fusion protein from E.coli.
Maciek GGS TEAM
Saturday, 4 December 2010
GGS LIVE - Foci kinetics
Yo, Yo, Yo Biofreak readers!!
Method: Foci kinetics.
About: Foci kinetics assay is performed in order to assess foci formation and resolution of particular protein under specific conditions.
What: Kinetics of gamma-H2AX foci formation after ionizing radiation in wild-type and mutant cells.
In our study case experiment we will investigate gamma-H2AX (histone modification) foci formation and resolution after treatment with ionizing radiation. Ionizing radiation cause DNA double strand breaks.
In response to such DNA damage H2AX histone is phosphorylated (called gamma-H2AX in phosphorylated state) to facilitate double strand break repair.
Briefly, DT40 cells were treated with IR (5 Gy), harvested and analysed by immunoflurescence at different times post-IR treatment (for immunofluorescence tutorial visit GGS LIVE - immunofluorescence). Pictures below represent cells stained for gamma-H2AX harvested at different times post-IR treatment.
As you see on the images above there is not much of signal from gamma-H2AX in untreated cells (there is small number of spontaneous DNA damage in unchallanged cells - red arrow heads indicate representative gamma-H2AX foci). After induction of double strand breaks with IR treatment H2AX histone is robustly phosphorylated (15 min timepoint) and this modification is removed with ongoing DNA repair. At this stage we have to quantify our results. We can eitehr simply score number of the foci per cell or score number of cells with more than X foci of gamma-H2AX. Plot below represents such quantification in which cells with 6 or more gamma-H2AX foci were scored as positive.
Method: Foci kinetics.
About: Foci kinetics assay is performed in order to assess foci formation and resolution of particular protein under specific conditions.
What: Kinetics of gamma-H2AX foci formation after ionizing radiation in wild-type and mutant cells.
In our study case experiment we will investigate gamma-H2AX (histone modification) foci formation and resolution after treatment with ionizing radiation. Ionizing radiation cause DNA double strand breaks.
In response to such DNA damage H2AX histone is phosphorylated (called gamma-H2AX in phosphorylated state) to facilitate double strand break repair.
Briefly, DT40 cells were treated with IR (5 Gy), harvested and analysed by immunoflurescence at different times post-IR treatment (for immunofluorescence tutorial visit GGS LIVE - immunofluorescence). Pictures below represent cells stained for gamma-H2AX harvested at different times post-IR treatment.
As you see on the images above there is not much of signal from gamma-H2AX in untreated cells (there is small number of spontaneous DNA damage in unchallanged cells - red arrow heads indicate representative gamma-H2AX foci). After induction of double strand breaks with IR treatment H2AX histone is robustly phosphorylated (15 min timepoint) and this modification is removed with ongoing DNA repair. At this stage we have to quantify our results. We can eitehr simply score number of the foci per cell or score number of cells with more than X foci of gamma-H2AX. Plot below represents such quantification in which cells with 6 or more gamma-H2AX foci were scored as positive.
From the quantification plot you can see that both cell lines induce gamma-H2AX foci fomration with the same kinetics, indicating that this process is not affected in the mutant cell line. When we look at gamma-H2AX foci resolution, four hours post-IR treatment 50% of wild-type cells has resolved gamm-H2AX foci, where mutant cells need another 4 hours to accomplish the same task. Such results indicate that mutant cell line might have problems with repairing DNA damage caused by IR treatment.
I hope you enjoyed:)
Maciek
GGS TEAM
Friday, 26 November 2010
GGS LIVE - Immunofluorescence
Yo Biofreakers:)
Today in the GGS LIVE section we are going to look at the immunofluorescence staining technique.
Method: Immunofluorescence.
About: Immunofluorecence is a technique that allows for detection of a particular protein within the cell.
What: Staining for gamma-H2AX and Rad51 proteins in chicken DT40 cells after ionizing radiation treatment.
Immunofluorescence technique is perfect for studying protein localization. Staining procedure is not compilcated and a representative protocol is shown on the cartoon below:
Briefly, DT40 suspension cells are harvested by centrifugation. Media is discarded and cell pellet is resuspended in 1 x PBS (phosphate buffered saline). Cells are adhered to poly-L-lysine slide, fixed and permeablised to allow access of molecules used in subsequent steps. Next, slide is incubated in blocking solution (usually a neutral protein, like 1 % bovine serum albumin solution). Blocking protein binds to sticky spots on slide, cells and within cells in order to decrease unspecific binding of the primiray and secondary antibodies (reduce unwanted background signal). After blocking, slides are incubated with primary antibodies that recognises the protein of interest. Notice that primary antibody has higher affinity to protein of interest and displace blocking protein. Slides are then washed to remove unspecifically bound antibodies. Later, secondary antibodies conjugated to a fluorophore are added and again washes are performed to remove unspecifically bound antibodies. Complex protein of interest - primary antibody - secondary antibody is formed and protein is ready to be detected and analysed by microscopy.
Lets have a look at our results.
As you can see in our study case experiment, cells treated with ionizing radiation were stained against two different proteins. Gamma-H2AX is a phosphorylated form of H2AX histone. H2AX histone is phosphorylated in proximity of DNA double strand break, appears as fast as 1-2 min after radiation and persists few hours post-treatment. Rad51 is a DNA repair protein and it accumulates at site of damage 2-4 hours after IR treatment. As you can see from our results both gamm-H2AX and Rad51 proteins form foci within nucleus (representative foci are indicated with yellow arrowheads). Both proteins nicely colocalise as seen on the overlay picture. With such immunofluorescence staining we can follow foci formation, resolution, change in protein localizatio or etc.
I hope you enjoyed:)
Maciek
GGS TEAM
Today in the GGS LIVE section we are going to look at the immunofluorescence staining technique.
Method: Immunofluorescence.
About: Immunofluorecence is a technique that allows for detection of a particular protein within the cell.
What: Staining for gamma-H2AX and Rad51 proteins in chicken DT40 cells after ionizing radiation treatment.
Immunofluorescence technique is perfect for studying protein localization. Staining procedure is not compilcated and a representative protocol is shown on the cartoon below:
Briefly, DT40 suspension cells are harvested by centrifugation. Media is discarded and cell pellet is resuspended in 1 x PBS (phosphate buffered saline). Cells are adhered to poly-L-lysine slide, fixed and permeablised to allow access of molecules used in subsequent steps. Next, slide is incubated in blocking solution (usually a neutral protein, like 1 % bovine serum albumin solution). Blocking protein binds to sticky spots on slide, cells and within cells in order to decrease unspecific binding of the primiray and secondary antibodies (reduce unwanted background signal). After blocking, slides are incubated with primary antibodies that recognises the protein of interest. Notice that primary antibody has higher affinity to protein of interest and displace blocking protein. Slides are then washed to remove unspecifically bound antibodies. Later, secondary antibodies conjugated to a fluorophore are added and again washes are performed to remove unspecifically bound antibodies. Complex protein of interest - primary antibody - secondary antibody is formed and protein is ready to be detected and analysed by microscopy.
Lets have a look at our results.
As you can see in our study case experiment, cells treated with ionizing radiation were stained against two different proteins. Gamma-H2AX is a phosphorylated form of H2AX histone. H2AX histone is phosphorylated in proximity of DNA double strand break, appears as fast as 1-2 min after radiation and persists few hours post-treatment. Rad51 is a DNA repair protein and it accumulates at site of damage 2-4 hours after IR treatment. As you can see from our results both gamm-H2AX and Rad51 proteins form foci within nucleus (representative foci are indicated with yellow arrowheads). Both proteins nicely colocalise as seen on the overlay picture. With such immunofluorescence staining we can follow foci formation, resolution, change in protein localizatio or etc.
I hope you enjoyed:)
Maciek
GGS TEAM
Tuesday, 23 November 2010
GGS LIVE - Cell fractionation
Method: Cellular fractionation.
About: Cellular fractionation is a technique that allows analysing protein localization to different cell compartments.
What: Comparison of protein X and Y localization in wild-type and mutant cell lines.
There are many protocols for cellular fractionation. Selection of a particular fractionation protocol may depend on cell type used, required fractionation resolution or protein analysed. In our study case we will look at a very basic fractionation protocol for mammallian cells. Cartoon below represents majors steps of the protocol:
Briefly, cells are incubated in hypothonic buffer to facilitate cell lysis. Cell membrane of swollen cells is mechanically disrupted by action of dounce homogeniser. At this stage cellular organelles are spun and supernatant containing cytoplasmic proteins is saved. Nuclei are incubated (usually with rocking or rotating) in high salt and detergent containing buffer to extract nuclear proteins. Again cell remaining are spun and supernatant containing nuclear proteins is saved. In the last step DNA bound proteins are extracted either by sonication (DNA sharing) or DNAase treatment (DNA digestion). After solubilization of DNA bound proteins extracts are spun and supernatant containing DNA bound proteins saved. All obtained samples can be now analysed by a method of choice. In our study case we will analyse our samples by Western blotting. Equal volume of each sample is separeted by SDS-PAGE to retaing similar loading and proteins are detected by Western blotting (for Western Blot tutorial click GGS LIVE Western Blotting). Ok, lets have a look at our results. In our experiment we have fractionated wild-type and mutant cell lines. To be sure that protocol worked perfectly, we have to first look at proteins that are known constituents of each cellular fraction. Cytoplasmic protein beta-actin, DNA binding protein Histone H3 and nuclear protein nucleophosmin are only present in cytoplasm, chromatin and nuclear fractions, respectively. Additionally all control proteins are present in Whole Cell Lysate sample. This is an additionall control which tells us if our protein of interest was present in the starting material.
As you can see analysis of protein X and Y reveals their distinct localization between wild-type and mutant cell line. When Protein X is exclusively cytoplasmic within wild-type cell line, it is also present in the nucleus of the mutant cell line. On the other hand protein Y is nuclear and partially bound to DNA in wild-type but its exclusively cytoplasmic in mutant cell line.
I hope you enjoyed it.
Maciek
GGS TEAM
About: Cellular fractionation is a technique that allows analysing protein localization to different cell compartments.
What: Comparison of protein X and Y localization in wild-type and mutant cell lines.
There are many protocols for cellular fractionation. Selection of a particular fractionation protocol may depend on cell type used, required fractionation resolution or protein analysed. In our study case we will look at a very basic fractionation protocol for mammallian cells. Cartoon below represents majors steps of the protocol:
Briefly, cells are incubated in hypothonic buffer to facilitate cell lysis. Cell membrane of swollen cells is mechanically disrupted by action of dounce homogeniser. At this stage cellular organelles are spun and supernatant containing cytoplasmic proteins is saved. Nuclei are incubated (usually with rocking or rotating) in high salt and detergent containing buffer to extract nuclear proteins. Again cell remaining are spun and supernatant containing nuclear proteins is saved. In the last step DNA bound proteins are extracted either by sonication (DNA sharing) or DNAase treatment (DNA digestion). After solubilization of DNA bound proteins extracts are spun and supernatant containing DNA bound proteins saved. All obtained samples can be now analysed by a method of choice. In our study case we will analyse our samples by Western blotting. Equal volume of each sample is separeted by SDS-PAGE to retaing similar loading and proteins are detected by Western blotting (for Western Blot tutorial click GGS LIVE Western Blotting). Ok, lets have a look at our results. In our experiment we have fractionated wild-type and mutant cell lines. To be sure that protocol worked perfectly, we have to first look at proteins that are known constituents of each cellular fraction. Cytoplasmic protein beta-actin, DNA binding protein Histone H3 and nuclear protein nucleophosmin are only present in cytoplasm, chromatin and nuclear fractions, respectively. Additionally all control proteins are present in Whole Cell Lysate sample. This is an additionall control which tells us if our protein of interest was present in the starting material.
As you can see analysis of protein X and Y reveals their distinct localization between wild-type and mutant cell line. When Protein X is exclusively cytoplasmic within wild-type cell line, it is also present in the nucleus of the mutant cell line. On the other hand protein Y is nuclear and partially bound to DNA in wild-type but its exclusively cytoplasmic in mutant cell line.
I hope you enjoyed it.
Maciek
GGS TEAM
Wednesday, 17 November 2010
GGS LIVE - Making a fusion protein
Yo BioFreakers,
today in the GGS LIVE section we will learn how to virtually design and generate a fusion protein.
Method: protein tagging.
About: protein tagging allows to perform specific experimenents that are not possible with the endogenous protein (wild-type protein).
What: desing and generation of protein X fusion with a GFP (green fluorescent protein) tag.
There are many different tags available for protein tagging. Choosing the tag depends on the experiments that we want to perfrorm with the fusion protein. So, if we want to:
- purify the protein of interest, we would use MBP (maltose binding protein), GST (gluthatione S-transferase), FLAG or His (hexahisitidine) tags,
- easily detect our protein, we would use epitope tags like myc, V5 or HA,
- follow cellular localization of the protein, we would use fluorescent tag like GFP.
The most important is to remember that fusion protein might behave differently in vivo than wild-type protein (folidng, solubility or activity, etc may change) so it is crucial to check if the fusion is functional before we start our experiments.
Ok lets start with design of the fusion protein. In our study case we will use a sequence of a protein X (shown below) and a pEGFP-C1 and -N1 plasmids for N and C terminal tagging, respectively.
The red triplets are start and stop codons, respectively. We are also going to need information about multi cloning site (MCS) sequence of the pEGFPN1 and -C1 vectors, where DNA sequence of protein X will be inserted. Information about those is shown below.
As you see there are many restriction sites available in both plasmids. Our aim here is to find a restriction enzymes that will cut in MCS of our vectors but not in the sequence of protein X (we can do that with any cloning software, like free pDRAW32 which you can download from here). In our study case two enzymes XhoI and EcoRI are cuttining in the MCSs but not in sequence of protein X. What we have to do now is to virtually introduce XhoI and EcoRI sequences at 5' and 3' end, respectively (sequences recognised by XhoI and EcoRI endonucleases are available here XhoI and EcoRI). There is one more issue to look at before we are going to add our restriction sites (see picture below).
It is important to remove start codon of the protein when tagging it on the N-terminus to avoid an expression of untagged form of the protein. You have to remember that promoter will drive expression of any open reading frame that is downstream of it. It is also crucial to remove stop codon when we tagging protein on the C-terminus to prevent premature termination and allow expression of a fusion protein. Including above information we have:
Now we put this sequence into our plasmid and we get this:
Lets have a look at our constructs now. We are going to focus on the reading frame at the moment. In the case of N-terminal tagging our reading frame is determined by the two last codons of GFP tag. To get the right fusion DNA sequence of protein X has to be in the reading frame with the GFP tag. The reading frame is indicated by the horizontal brackets (each triplet codes for one amino acid). As you can see stop codon of the protein X (indicated with the red colour) is not in reading frame with GFP tag. To shift the reading frame we have to add two extra nuclotides to our protein X just between the XhoI restriction site and the coding sequence of protein X. Similar situation takes place with the C-terminal tagging. You can see that now start codon is not in frame with GFP tag and addition of a single nuclotide between EcoRI restriction site and protein sequence will rescue that problem (see the pictures below).
It is important to remember that addition of extra nuclotides to our sequence may result in introduction of the stop codon. We have to check our sequence before we proceed further. If everything is ready we should obtain this:
As you can see now, protein X sequence is in the frame with the GFP tag in both cases. Now the protein sequence including the XhoI / EcoRI restriction sites and extra nucleotides can be used to design primers for cloning of the protein X DNA. Such DNA then will be sequenced to check for potential mutations and if correct subcloned into pEGFPN1 or -C1 plasmids. Such construct can be later used for expression and localization studies of the protein X.
I hope u enjoyed it.
Maciek
GGS TEAM
today in the GGS LIVE section we will learn how to virtually design and generate a fusion protein.
Method: protein tagging.
About: protein tagging allows to perform specific experimenents that are not possible with the endogenous protein (wild-type protein).
What: desing and generation of protein X fusion with a GFP (green fluorescent protein) tag.
There are many different tags available for protein tagging. Choosing the tag depends on the experiments that we want to perfrorm with the fusion protein. So, if we want to:
- purify the protein of interest, we would use MBP (maltose binding protein), GST (gluthatione S-transferase), FLAG or His (hexahisitidine) tags,
- easily detect our protein, we would use epitope tags like myc, V5 or HA,
- follow cellular localization of the protein, we would use fluorescent tag like GFP.
The most important is to remember that fusion protein might behave differently in vivo than wild-type protein (folidng, solubility or activity, etc may change) so it is crucial to check if the fusion is functional before we start our experiments.
Ok lets start with design of the fusion protein. In our study case we will use a sequence of a protein X (shown below) and a pEGFP-C1 and -N1 plasmids for N and C terminal tagging, respectively.
The red triplets are start and stop codons, respectively. We are also going to need information about multi cloning site (MCS) sequence of the pEGFPN1 and -C1 vectors, where DNA sequence of protein X will be inserted. Information about those is shown below.
As you see there are many restriction sites available in both plasmids. Our aim here is to find a restriction enzymes that will cut in MCS of our vectors but not in the sequence of protein X (we can do that with any cloning software, like free pDRAW32 which you can download from here). In our study case two enzymes XhoI and EcoRI are cuttining in the MCSs but not in sequence of protein X. What we have to do now is to virtually introduce XhoI and EcoRI sequences at 5' and 3' end, respectively (sequences recognised by XhoI and EcoRI endonucleases are available here XhoI and EcoRI). There is one more issue to look at before we are going to add our restriction sites (see picture below).
It is important to remove start codon of the protein when tagging it on the N-terminus to avoid an expression of untagged form of the protein. You have to remember that promoter will drive expression of any open reading frame that is downstream of it. It is also crucial to remove stop codon when we tagging protein on the C-terminus to prevent premature termination and allow expression of a fusion protein. Including above information we have:
Now we put this sequence into our plasmid and we get this:
Lets have a look at our constructs now. We are going to focus on the reading frame at the moment. In the case of N-terminal tagging our reading frame is determined by the two last codons of GFP tag. To get the right fusion DNA sequence of protein X has to be in the reading frame with the GFP tag. The reading frame is indicated by the horizontal brackets (each triplet codes for one amino acid). As you can see stop codon of the protein X (indicated with the red colour) is not in reading frame with GFP tag. To shift the reading frame we have to add two extra nuclotides to our protein X just between the XhoI restriction site and the coding sequence of protein X. Similar situation takes place with the C-terminal tagging. You can see that now start codon is not in frame with GFP tag and addition of a single nuclotide between EcoRI restriction site and protein sequence will rescue that problem (see the pictures below).
It is important to remember that addition of extra nuclotides to our sequence may result in introduction of the stop codon. We have to check our sequence before we proceed further. If everything is ready we should obtain this:
As you can see now, protein X sequence is in the frame with the GFP tag in both cases. Now the protein sequence including the XhoI / EcoRI restriction sites and extra nucleotides can be used to design primers for cloning of the protein X DNA. Such DNA then will be sequenced to check for potential mutations and if correct subcloned into pEGFPN1 or -C1 plasmids. Such construct can be later used for expression and localization studies of the protein X.
I hope u enjoyed it.
Maciek
GGS TEAM
Friday, 5 November 2010
GGS LIVE - Immuno and co-immunoprecipitation
Whatsup you all?
GGS LIVE sections presents today an immunoprecipitation method:)
Method: Immunoprecipitation.
About: Protein precipitation using specific antibodies is a technique that allows for enrichment of a particular protein and components associated with this protein (it is a type of protein purification).
What: Immunoprecipitation using anti-myc antibody from DT40 cells stably expressing 3myc tagged protein.
This technique can be used to purify the protein of interest in order to use it in a specific assay or to identify its potential interacting partners. For example an enzyme protein kinase can be immunoprecipitated and subsequently used in the kinase assay. The cartoon below show immunoprecipitation steps.
GGS LIVE sections presents today an immunoprecipitation method:)
Method: Immunoprecipitation.
About: Protein precipitation using specific antibodies is a technique that allows for enrichment of a particular protein and components associated with this protein (it is a type of protein purification).
What: Immunoprecipitation using anti-myc antibody from DT40 cells stably expressing 3myc tagged protein.
This technique can be used to purify the protein of interest in order to use it in a specific assay or to identify its potential interacting partners. For example an enzyme protein kinase can be immunoprecipitated and subsequently used in the kinase assay. The cartoon below show immunoprecipitation steps.
Briefly, cell lysate is prepared by cell lysis in IP buffer of choice (the composition of the buffer might be different from protein to protein analysed and may have to be found empirically). Cell lysate is also sonicated to share the DNA that may interfere with the immunoprecipitation process and also to solubilize proteins. Next, cell membrane and debri is spun down and protein concentration is determined. On this stage the lysate is ready to go. We also should prepare our beads and antibody. The Protein A or G beads are washed three times with lysis buffer (aka IP buffer) to remove beads storage solution (usually ethanol which can interfere with the precedure). Washed beads are mixed with antibodies. After coupling, beads are mixed with previously prepared cell lysate and this is our immunoprecipitation step. After precipitation beads + antibodies + protein of interest + partner proteins are washed with lysis buffer (or other buffers) to remove unspecifically bound material. Finally, precipitated proteins are eluted and analysed by Western blotting (for the Western blotting tutorial visit this post GGS LIVE - Western Blotting). Ok lets see the results now...
On the top panel we see bands representing the protein against which the antibody was used (immunoprecipitated protein). Bottom panel shows co-precipitated protein (a partner protein of the myc-tagged protein of interest). The lanes are: input lane, representing starting material, unbound lane showing material that left after the immunoprecipitation and finally the IP lane which represents the precipitated proteins fraction. The control IP has to be performed so we are sure that the protein we are enriching for is specifically pulled down. One can imagine that protein of interest might bind to other part of the antibody (that is common for anti-myc or control antibody) or bind unspecifcally for example to the beads. Simply saying the signal form our protein should be present only in the experiment IP but not in the control IP. As you can see this is correct for our myc tagged protein which is present in the starting material, it is depleted in unbound fraction and there is an obvious enrichment of it on the beads (IP lane). In the case of the control IP again it is present in the input lane but it is absent in the IP lane (which is perfect:).
The same samples were analysed for the presence of a partner protein that is known to interact with our myc-tagged target. You can clearly see that the interacting partner is also present in all the samples beside the control IP what suggests a that it is interacting with the myc-tagged protein. Of course novel interactions have to be confirmed with reciprocal experiment.
I hope u enjoyed and understand that immunoprecipitation is a very powerful cell biology technique:)
Maciek
GGS TEAM
Tuesday, 2 November 2010
GGS LIVE - How to measure DNA synthesis in vivo?
Hello Biofreakers,
Today in the GGS LIVE section we are going to look at techniques that allow to observe cells that replicate DNA (are in the S-phase of the cell cycle).
Method: Labelling and detection of replicating (aka. S-phase) cells.
About: Both methods that will be discussed (flow cythometry and immunofluorescence) use specific derivative of nucleotide that is incorporated into DNA during the S-phase of cell cycle. Later this modified nuclotide can be detected and S-phase cells analysed.
What: Labelling of chicken DT40 S-phase cells.
One of the most common nucleotide derivative used in cell biology is a bromodeoxyuridine (BrdU, for structure and other applications go here Meet the chickens - sister chromatid labelling). BrdU pulse will result in its incorporation into cells DNA. In our case study such cells will be analysed by flow cytometry and immunofluorescence. This techniques are very different but sample preparation is very similar, see picture below.
And this one:)
Similarly like in the Western and Southern Blotting, antibody used here is conjugated to a moiety that gives a specific signal, in this case antibody is conjugated to a green fluorophore called FITC. The signal from fluorophore can be detected in many different ways. In our study case, cells designated for flow cytometry will be analysed with flow cytometer:) and ones on the slide will be analysed with fluorescent microscope. Ok lets check the results of our experiment.
Lets look first at the panel A where S-phase labelled cells are analysed by flow cytometry (for those that are not familiar with flow cytometry, please visit this post GGS LIVE - flow cytometry). The first histogram in the panel A represents a DNA content profile. On the x-axis we have a DNA content. The first big peak are cells in G1 phase of the cell cyle with 2N DNA content, second smaller peak are cells in G2/M phase of cell cycle with 4N DNA content and finally all cells between those two peaks are cells in S-phase with DNA content between 2N-4N. On the y-axis we have counts (the BrdU incporporation is only relevant for the other two dot plots). So you can easily see that the most number of the cells is in the G1 phase of the cell cycle followed by S-phase and G2/M cells. This histogram is often called one dimensional cell cycle plot as it measure only DNA content. The other two dot plots are often called two dimensional as we measure DNA content and BrdU incorporation. What you can see on the first dot plot is exatly the same as on the first histogram but represented in a slightly different way. Now, there is no peaks but each dot represents a single cell. Similarly the first bunch of dots (cells:) is population of G1 phase cells and the other bunch is population of G2/M cells. Everyhting between them are S-phase cells. Look know on the last dot plot. The situation is similar but these cells were pulsed with BrdU and what you can see know is that S-phase cells did shiftet to upper values on the y-axis (which is a measure of the fluorescence that is directly proportional to the antibody that recognise BrdU). This picture is often called a horseshoe.
On the panel B we can see pictures of cells. On the first panel we see DNA that was labelled with DAPI (blue flurophore), in the middle we see a replication foci that were lebelled with BrdU and detected with antibodies. On the last picture we see an overaly of the two, which clearly indicates that replication foci are localised to DNA (what makes sense:).
Both techniques are very useful in analysis of replicating cells. The flow cytometry allows for a very robust and quick analysis of S-phase cells (be aware that flow cytometry allows for quantification of the cells in each phase of cell cycle). We can use this technique to monitor S-phase cells for example after treatment with different drugs. Immunofluorecence method does not have power of numbers but allows detecting of for example other proteins colocalization to replication foci (if protein X will be labelled red we can see if it colocalizes with replication foci). Both techniques are very powerful and are commonly used in each cell biology laboratory.
I hope u enjoyed:)
Maciek
GGS TEAM
Today in the GGS LIVE section we are going to look at techniques that allow to observe cells that replicate DNA (are in the S-phase of the cell cycle).
Method: Labelling and detection of replicating (aka. S-phase) cells.
About: Both methods that will be discussed (flow cythometry and immunofluorescence) use specific derivative of nucleotide that is incorporated into DNA during the S-phase of cell cycle. Later this modified nuclotide can be detected and S-phase cells analysed.
What: Labelling of chicken DT40 S-phase cells.
One of the most common nucleotide derivative used in cell biology is a bromodeoxyuridine (BrdU, for structure and other applications go here Meet the chickens - sister chromatid labelling). BrdU pulse will result in its incorporation into cells DNA. In our case study such cells will be analysed by flow cytometry and immunofluorescence. This techniques are very different but sample preparation is very similar, see picture below.
And this one:)
Similarly like in the Western and Southern Blotting, antibody used here is conjugated to a moiety that gives a specific signal, in this case antibody is conjugated to a green fluorophore called FITC. The signal from fluorophore can be detected in many different ways. In our study case, cells designated for flow cytometry will be analysed with flow cytometer:) and ones on the slide will be analysed with fluorescent microscope. Ok lets check the results of our experiment.
Lets look first at the panel A where S-phase labelled cells are analysed by flow cytometry (for those that are not familiar with flow cytometry, please visit this post GGS LIVE - flow cytometry). The first histogram in the panel A represents a DNA content profile. On the x-axis we have a DNA content. The first big peak are cells in G1 phase of the cell cyle with 2N DNA content, second smaller peak are cells in G2/M phase of cell cycle with 4N DNA content and finally all cells between those two peaks are cells in S-phase with DNA content between 2N-4N. On the y-axis we have counts (the BrdU incporporation is only relevant for the other two dot plots). So you can easily see that the most number of the cells is in the G1 phase of the cell cycle followed by S-phase and G2/M cells. This histogram is often called one dimensional cell cycle plot as it measure only DNA content. The other two dot plots are often called two dimensional as we measure DNA content and BrdU incorporation. What you can see on the first dot plot is exatly the same as on the first histogram but represented in a slightly different way. Now, there is no peaks but each dot represents a single cell. Similarly the first bunch of dots (cells:) is population of G1 phase cells and the other bunch is population of G2/M cells. Everyhting between them are S-phase cells. Look know on the last dot plot. The situation is similar but these cells were pulsed with BrdU and what you can see know is that S-phase cells did shiftet to upper values on the y-axis (which is a measure of the fluorescence that is directly proportional to the antibody that recognise BrdU). This picture is often called a horseshoe.
On the panel B we can see pictures of cells. On the first panel we see DNA that was labelled with DAPI (blue flurophore), in the middle we see a replication foci that were lebelled with BrdU and detected with antibodies. On the last picture we see an overaly of the two, which clearly indicates that replication foci are localised to DNA (what makes sense:).
Both techniques are very useful in analysis of replicating cells. The flow cytometry allows for a very robust and quick analysis of S-phase cells (be aware that flow cytometry allows for quantification of the cells in each phase of cell cycle). We can use this technique to monitor S-phase cells for example after treatment with different drugs. Immunofluorecence method does not have power of numbers but allows detecting of for example other proteins colocalization to replication foci (if protein X will be labelled red we can see if it colocalizes with replication foci). Both techniques are very powerful and are commonly used in each cell biology laboratory.
I hope u enjoyed:)
Maciek
GGS TEAM
Tuesday, 14 September 2010
GGS LIVE - Counting cells
Hello BioFreaks Reader,
Today in GGS LIVE section we are having some basic Tissue Culture technique:
Method: Determination of cell number or cell density in the culture.
About: Determination of cell number is essential in performing experiments where appropriate number of cells is required in order to sustain uniform conditions during the experiment.
What: Determination of cell number in suspension cell culture.
The most common technique to determine cell number in culture is to count single cells using a microscope and a hemacytometer (see picture below).
To count suspension cells we need to first mixed them well to homogenise the entire culture. After mixing 10microliteres is taken and loaded onto a hemocytometer as shown on pictures below.
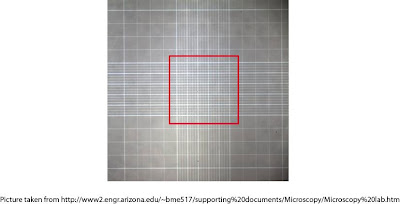
To count cells using hemocytometer we need to hava a closer look at it. Under magnification smooth area of hemocytometer looks like this:
The lilquid that occupy area of a centre square (here indicated with red border) which contains 25 smaller squares has volume of 0.1microliter. After loading cell onto hemocytometer we should see something like this:
The red boarder again indicates the central square that we are going to score. Additionally red arrowheads inducate three represenatative cells. We should count cells at least twice and take an average of two as the final result. For example: count one 93 cells
count two 87 cells
total 180 cells
average 90 cells
To get a number of cells in a 1ml of culture we need to multiply our result by 10 000. Why? because volume scored is 0.1microliter and that is 10 000 less tham 1ml, so in 1ml there is 10 000 more cells than in the volume we had investigated (if you have problem with prefixes and calculations visit this post Scientific prefixes - you will be laughing). So in our study case we have 90 x 10 000 = 900 000 cells in 1ml.
I hope you enjoyed it.
Maciek GGSTEAM
Today in GGS LIVE section we are having some basic Tissue Culture technique:
Method: Determination of cell number or cell density in the culture.
About: Determination of cell number is essential in performing experiments where appropriate number of cells is required in order to sustain uniform conditions during the experiment.
What: Determination of cell number in suspension cell culture.
The most common technique to determine cell number in culture is to count single cells using a microscope and a hemacytometer (see picture below).
To count suspension cells we need to first mixed them well to homogenise the entire culture. After mixing 10microliteres is taken and loaded onto a hemocytometer as shown on pictures below.
To count cells using hemocytometer we need to hava a closer look at it. Under magnification smooth area of hemocytometer looks like this:
The lilquid that occupy area of a centre square (here indicated with red border) which contains 25 smaller squares has volume of 0.1microliter. After loading cell onto hemocytometer we should see something like this:
The red boarder again indicates the central square that we are going to score. Additionally red arrowheads inducate three represenatative cells. We should count cells at least twice and take an average of two as the final result. For example: count one 93 cells
count two 87 cells
total 180 cells
average 90 cells
To get a number of cells in a 1ml of culture we need to multiply our result by 10 000. Why? because volume scored is 0.1microliter and that is 10 000 less tham 1ml, so in 1ml there is 10 000 more cells than in the volume we had investigated (if you have problem with prefixes and calculations visit this post Scientific prefixes - you will be laughing). So in our study case we have 90 x 10 000 = 900 000 cells in 1ml.
I hope you enjoyed it.
Maciek GGSTEAM
Monday, 13 September 2010
GGS LIVE - Clonogenic Survival Assay
Yo Nerds,
Method: Clonogenic Survival Assay (CSA).
About: Clonogenic survival assay is a long term cell viability assay in which ability of a single cell to form a colony (proliferation capacity) is scored.
What: Clonogenic survival assay after DNA damage induced by Drug X using two different cell lines: wild-type and DNA repair defficient mutant.
Clonogenic survival assay is performed differently for suspension and adherent cells. As I am working with suspension cell at the moment, I will explain only how to perform CSA for suspension cells.
Around 5 x 10^5 cells is enough to perform clonogenic survival assay with four different doses of Drug X.
If we perform clonogenic survival assay for the first time using a mutant cell line (or using a particular drug for the first time) it is important to plate different number of cells for each dose as it is hard to predict sensitivity, for example:
It is also important to seed cells in triplicate. This is because we want to base our result on a three different numbers what makes it more accurate (not on a single number but on avarage of three). When we have our cells and numbers ready we have to prepare plates. Different plates can be used for this purpose but in our lab we are using the triple vent petri dishes (see on the picture further on).
Special media has to be used in the case of suspension cells, in our lab we are using a methylcellulose based media. Methylcellulose is added to the media as a thickener so our "mobile" suspension cells do not move after we plate them. This prevents a single cell to give rise to multiple colonies. You can imagine that if media is not thick colony could split resulting in extra number of colonies at the end of the experiment. Once the media is placed in dishes we can start plating cells. For that purpose a dilution method is used, as show on the picture below:
We prepare our cells so we have a 10^5 cells per ml and we place them in 24 well plate (picture above).
We treat this cells with Drug X at doses 0, 2, 4 and 8 mg/ml (in the 0 dose we treat cells with the solvent used to dissolve the Drug X only). We incubate the cells at 37 degrees Celsius for specific time and after that time we dilute cells twice each time by factor of 10 (as on the picture above). This way we obtain three cell solutions in which we have 100 000, 10 000 and 1000 cells in total per ml. This means that if we use 100ul from each solution we will plate 10 000, 1000 and 100 cells onto dish, respectively. After the cells are plated we incubate them for 7-14 days depending on how fast cells grow and give visible colonies (see picture below).
Three representative colonies are indicated with red arrowheads. We count colonies and then we calculate the survival according to the table shown below:
After the calculations we plot the % of relative survival against the doses of Drug X and we obtain a plot like on the picture below:
From this experiment we can conclude that mutant cell line is more sensitive to Drug X than wild-type cells and that this sensitivity is dose dependent. We have to repeat this experiment at least three times to obtain reliable result. Remember that all experimental steps (cell number, drug doses, time of cell treatment) has to be optimized and each time performed in the same way to minimize the errors.
I hope u enjoyed it:)
Maciek GGSTEAM
Method: Clonogenic Survival Assay (CSA).
About: Clonogenic survival assay is a long term cell viability assay in which ability of a single cell to form a colony (proliferation capacity) is scored.
What: Clonogenic survival assay after DNA damage induced by Drug X using two different cell lines: wild-type and DNA repair defficient mutant.
Clonogenic survival assay is performed differently for suspension and adherent cells. As I am working with suspension cell at the moment, I will explain only how to perform CSA for suspension cells.
Around 5 x 10^5 cells is enough to perform clonogenic survival assay with four different doses of Drug X.
If we perform clonogenic survival assay for the first time using a mutant cell line (or using a particular drug for the first time) it is important to plate different number of cells for each dose as it is hard to predict sensitivity, for example:
It is also important to seed cells in triplicate. This is because we want to base our result on a three different numbers what makes it more accurate (not on a single number but on avarage of three). When we have our cells and numbers ready we have to prepare plates. Different plates can be used for this purpose but in our lab we are using the triple vent petri dishes (see on the picture further on).
Special media has to be used in the case of suspension cells, in our lab we are using a methylcellulose based media. Methylcellulose is added to the media as a thickener so our "mobile" suspension cells do not move after we plate them. This prevents a single cell to give rise to multiple colonies. You can imagine that if media is not thick colony could split resulting in extra number of colonies at the end of the experiment. Once the media is placed in dishes we can start plating cells. For that purpose a dilution method is used, as show on the picture below:
We prepare our cells so we have a 10^5 cells per ml and we place them in 24 well plate (picture above).
We treat this cells with Drug X at doses 0, 2, 4 and 8 mg/ml (in the 0 dose we treat cells with the solvent used to dissolve the Drug X only). We incubate the cells at 37 degrees Celsius for specific time and after that time we dilute cells twice each time by factor of 10 (as on the picture above). This way we obtain three cell solutions in which we have 100 000, 10 000 and 1000 cells in total per ml. This means that if we use 100ul from each solution we will plate 10 000, 1000 and 100 cells onto dish, respectively. After the cells are plated we incubate them for 7-14 days depending on how fast cells grow and give visible colonies (see picture below).
Picture taken by Kliszczak M.
Three representative colonies are indicated with red arrowheads. We count colonies and then we calculate the survival according to the table shown below:
After the calculations we plot the % of relative survival against the doses of Drug X and we obtain a plot like on the picture below:
From this experiment we can conclude that mutant cell line is more sensitive to Drug X than wild-type cells and that this sensitivity is dose dependent. We have to repeat this experiment at least three times to obtain reliable result. Remember that all experimental steps (cell number, drug doses, time of cell treatment) has to be optimized and each time performed in the same way to minimize the errors.
I hope u enjoyed it:)
Maciek GGSTEAM
Monday, 10 May 2010
GGS LIVE - Flow cytometry
Yo all,
welcome again in the Biochemistry Method section. Today we will cover a Flow Cytometry Technique.
Method: Flow cytometry
About: simply saying flow cytometry is a technique where properties of microscopic particles (such as cells or chromosomes) is examined. Flow cytometry is a very powerful method because it can measure many particles in the same time (up to thousands). Additionally, flow cytometry analysis is multiparametric, so different properties of particles can be measured at once.
What: Analysis of cell cycles distribution in different populations of cells.
How: By flow cytometry.
Ok, so let's start with some short background on how the flow cytometer looks like (see picture below)
and how it works ... (what is inside? :) picture below)
Of course this is a simplified scheme:). What you can see on the scheme is:
- a tube with your sample (in our case cells are the particles examined),
- sample collector,
- source of liquids,
- mirrors,
- laser,
- detectors,
- and waster container.
To simplify how the flow cytometer works we can say that particles are sucked from tube, enter machine flow, where they are exposed to laser and finish in waste container. Depending on particles they either adsorb or alter path of the light. This is monitored by different detectors installed in the flow cytometer. Two most important are:
- forward scatter detector - which is placed in line with the light beam and gives us information about the size of cells (particles),
- side scatter - which is placed perpendicularly to light beam and gives us information about surface of cells (particles) for example their roughness etc.
Additional detectors are installed in the flow cytometer which are able to detect other features of cells/particles (for example they can measure fluorescnce of a chemical compund bound to cell membrane giving us idea how many cells do contain such modified membrane).
In our case study we will look at cell DNA content. Ok let's start. To perform flow cytometry we need to first prepare cells. Our experimental design is as follow:
- one control sample (untreated cells, also called unsynchronous population),
- and three treated samples:
After short treatment (when complex between DNA and dye is formed) cells are ready for analysis. So, we simply kick in machine, install our samples and run analysis. The control result from our experiment is shown on picture below.
The left-hand plot is a representation of our population (below you can see it as indicated with a red area), where each spot is a single event (cell or particle). The X-axis is a forward scatter (FSC), which if you remember tells us about the size of particle. If we move along it we go from the smallest cells (G1 phase cells), through S-phase, to the biggest G2 and mitotic cells. The Y-axis is a side scatter (SSC) and if you remeber it tells us about the sufrace of particle/cell. As you can imagine cells in G1 phase have different morphology than cells in S-, G2 or M-phase (that's why they have different position on Y-axis). We can alter position of our population on the dot plot using specific parameters but usually we try to place it in diagonal of dot plot box and between value 200 and 400 on X-axis.
If you have a look at the histogram plot now, you can see that on X-axis we have FL2 parameter (which is actually a fluorescence of our DNA dye) and on Y-axis counts. This plot simply tells us how many cells contains how much of fluorescence. You can imagine that cell in G1 phase, where there is a single copy of DNA, will have smaller fluorescence than a G2 cell which contains doubled amount of DNA. G1 peak is placed at 200 and G2 peak at 400. Everything in between is S-phase cells which contain amount of DNA between 1n and 2n. From this you can see that the most number of cells are in G1 phase then in S- and in G2-phase.
Now let's have a look at flow cytometry results of our treated cells.
We have already covered control experiment, so we start from Nocodazole treated sample.
Nocodazole stops cells at G2/M boarded. You can easily see that dot plot and histogram plot shifted to the right. The block of cells might not be so clear at dot plot but from histogram we can tell that all cells have been stalled in G2 phase (peak at 400). There is no G1-peak or S-phase area.
Hydroxyurea which prevents cells to enter S-phase, decreasaed number of S-phase and G2/M cells. G1 peak is now fatter what suggest that most of cells is stalled in G1 phase. This block is not nice as Nocodazole one but I hope you see difference between hydroxyurea treated and control cells.
Drug X treatment is toxic to cells. We can deffinitely say that it dimnishes S-phase cells and block them in G1. Additionally you can see a small sub-G1 peak which is actually a indication of apoptotic cells (during apoptosis- a programmed cell death, DNA of cell is fragmented and distributed to apoptotic bodies. This is why it appears as smaller, less than 1n).
This is it:) I hope you enjoy it.
Maciek GGSTEAM
welcome again in the Biochemistry Method section. Today we will cover a Flow Cytometry Technique.
Method: Flow cytometry
About: simply saying flow cytometry is a technique where properties of microscopic particles (such as cells or chromosomes) is examined. Flow cytometry is a very powerful method because it can measure many particles in the same time (up to thousands). Additionally, flow cytometry analysis is multiparametric, so different properties of particles can be measured at once.
What: Analysis of cell cycles distribution in different populations of cells.
How: By flow cytometry.
Ok, so let's start with some short background on how the flow cytometer looks like (see picture below)
and how it works ... (what is inside? :) picture below)
Of course this is a simplified scheme:). What you can see on the scheme is:
- a tube with your sample (in our case cells are the particles examined),
- sample collector,
- source of liquids,
- mirrors,
- laser,
- detectors,
- and waster container.
To simplify how the flow cytometer works we can say that particles are sucked from tube, enter machine flow, where they are exposed to laser and finish in waste container. Depending on particles they either adsorb or alter path of the light. This is monitored by different detectors installed in the flow cytometer. Two most important are:
- forward scatter detector - which is placed in line with the light beam and gives us information about the size of cells (particles),
- side scatter - which is placed perpendicularly to light beam and gives us information about surface of cells (particles) for example their roughness etc.
Additional detectors are installed in the flow cytometer which are able to detect other features of cells/particles (for example they can measure fluorescnce of a chemical compund bound to cell membrane giving us idea how many cells do contain such modified membrane).
In our case study we will look at cell DNA content. Ok let's start. To perform flow cytometry we need to first prepare cells. Our experimental design is as follow:
- one control sample (untreated cells, also called unsynchronous population),
- and three treated samples:
- nocodazole treated cells (nocodazole is microtubule depolymerizing agent causing cells to stall at the G2/M boarded). For the cell cycle tutorial please visit this post Theory is fundamental - Cell cycle.
- hydroxy urea treated cells (HU- hydroxy urea ribonucleotide reductase inhibitor. Inhibition of this enzyme leads to depletion of DNA synthesis substrates - deoxyribonucleotides). This drug stalls cells in or before S-phase.
- Drug X treated cells.
After short treatment (when complex between DNA and dye is formed) cells are ready for analysis. So, we simply kick in machine, install our samples and run analysis. The control result from our experiment is shown on picture below.
The left-hand plot is a representation of our population (below you can see it as indicated with a red area), where each spot is a single event (cell or particle). The X-axis is a forward scatter (FSC), which if you remember tells us about the size of particle. If we move along it we go from the smallest cells (G1 phase cells), through S-phase, to the biggest G2 and mitotic cells. The Y-axis is a side scatter (SSC) and if you remeber it tells us about the sufrace of particle/cell. As you can imagine cells in G1 phase have different morphology than cells in S-, G2 or M-phase (that's why they have different position on Y-axis). We can alter position of our population on the dot plot using specific parameters but usually we try to place it in diagonal of dot plot box and between value 200 and 400 on X-axis.
If you have a look at the histogram plot now, you can see that on X-axis we have FL2 parameter (which is actually a fluorescence of our DNA dye) and on Y-axis counts. This plot simply tells us how many cells contains how much of fluorescence. You can imagine that cell in G1 phase, where there is a single copy of DNA, will have smaller fluorescence than a G2 cell which contains doubled amount of DNA. G1 peak is placed at 200 and G2 peak at 400. Everything in between is S-phase cells which contain amount of DNA between 1n and 2n. From this you can see that the most number of cells are in G1 phase then in S- and in G2-phase.
Now let's have a look at flow cytometry results of our treated cells.
We have already covered control experiment, so we start from Nocodazole treated sample.
Nocodazole stops cells at G2/M boarded. You can easily see that dot plot and histogram plot shifted to the right. The block of cells might not be so clear at dot plot but from histogram we can tell that all cells have been stalled in G2 phase (peak at 400). There is no G1-peak or S-phase area.
Hydroxyurea which prevents cells to enter S-phase, decreasaed number of S-phase and G2/M cells. G1 peak is now fatter what suggest that most of cells is stalled in G1 phase. This block is not nice as Nocodazole one but I hope you see difference between hydroxyurea treated and control cells.
Drug X treatment is toxic to cells. We can deffinitely say that it dimnishes S-phase cells and block them in G1. Additionally you can see a small sub-G1 peak which is actually a indication of apoptotic cells (during apoptosis- a programmed cell death, DNA of cell is fragmented and distributed to apoptotic bodies. This is why it appears as smaller, less than 1n).
This is it:) I hope you enjoy it.
Maciek GGSTEAM
Subscribe to:
Posts (Atom)